The implementation of DNA-based methods like polymerase chain reaction in water testing is a new concept for South Africa, and ERWAT was one of the first laboratories to implement these methods. Water&Sanitation Africa speaks to Nico van Blerk, microbiologist at ERWAT Laboratory Services, about this specialised facility.
The initiative to implement advanced technology to support the standard microbiological laboratory was taken in early 2007. This followed a water-related Salmonella outbreak, during which the ERWAT microbiology laboratory provided analytical services. Using standard culturing methods had several disadvantages. First, culturing methods are laborious and time consuming, requiring three to seven days to complete. Second, culturing methods are known to lack sensitivity and selectivity, making it difficult to successfully isolate and identify pathogens such as Salmonella and Vibrio cholerae, especially from sample matrices with a complex natural microbial population (e.g. river/dam water samples). Receiving many urgent samples, especially during the outbreak, the laboratory was overloaded – resulting in long hours of work. This led to the question whether there was any better and faster way to do pathogen detections and it was decided to establish a polymerase chain reaction (PCR) laboratory. In early 2012, the PCR laboratory was assessed by the South African National Accreditation System and three of the real-time PCR detection as says are now accredited. These are the detection of Salmonella enterica, Shigella species and/or entero-pathogenic E. coli, and toxigenic Vibrio cholerae (cholera causing). DNA-based detection methods – a powerful alternative The phenotypical characteristics of all living organisms, including bacteria, are locked up in their DNA. Therefore the genes in a particular species of bacteria are unique. This provides a useful marker to detect and identify the organism. PCR is a technique specifically designed to target and copy (amplify) genes. PCR is now a commonly used techniques in molecular biology laboratories – from research to forensic, medical and food laboratories. The advantages of using PCR detection assays instead of culturing assays are that PCR is highly selective for the pathogen targeted and much faster. Accurate results achieved in less than 24 hours.PCR basics
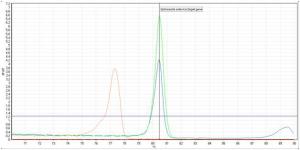
PCR employs a special enzyme (DNA polymerase) to incorporate the individual building blocks of DNA (A, T, G and G) onto a template of the targeted gene, creating an exact copy of that gene. Repeating this procedure 30 to 40 times results in millions of exact copies of the original gene, making it possible for detection and identification. During the past fiveyears, significant advancements regarding DNA binding dyes and PCR instrumentation now allow for the PCR process to be observed and monitored in real time. Real-time PCR is more sensitive and allows for post-amplification analyses of the gene copies such as quantification, melt curve and High Resolution Melt (HRM) curve. High Resolution Melt curve analysis DNA molecules consist of two strands bound to one another by hydrogen bonds. Using heat, these hydrogen bonds can be broken causing the separation of the two strands and subsequently the melting (denaturation) of the DNA molecule. The temperature required to melt 50% of a particular DNA product is known as the melting temperature (Tm) of that product. The Tm of a DNA product is related to its sequence.. Using this property of DNA molecules and specialised instrumentation to monitor the melting process, the identity of a product formed by PCR amplification can be determined. Thus, by comparing the Tm of an unknown PCR product to that of a known PCR product (i.e. a positive control), the identity of the unknown PCR product can be determined (as shown in Figure 1).